About
What is NERDSS?
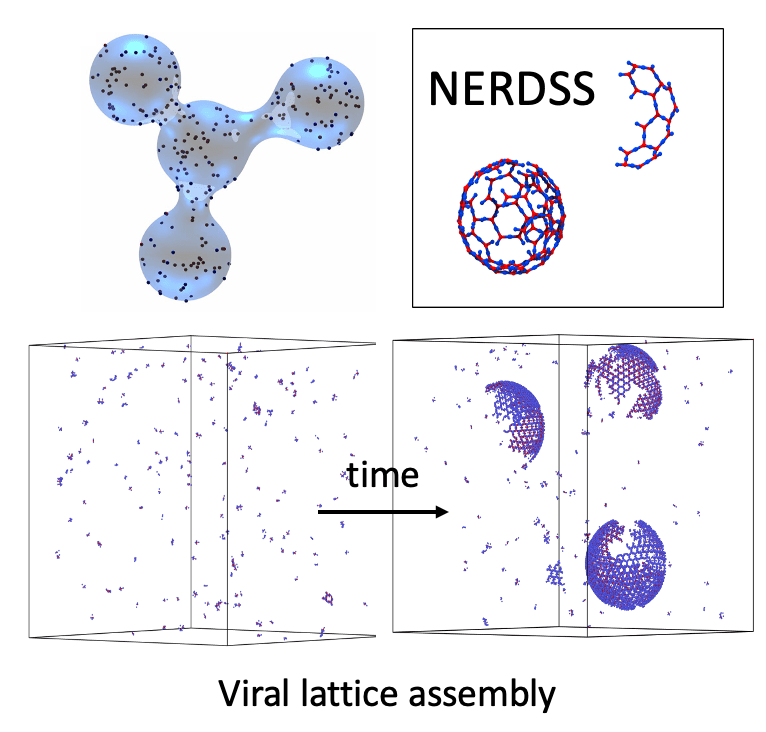
NERDSS, or non-equailbrium reaction-diffusion self-assembly simulator, was developed to build self-assembly into the reaction-diffusion (RD) model because of the strength of RD in simulating relatively slow, nonequilibrium dynamics. The name of the software may sound somewhat redundant (particularly NERD). The “nonequilibrium” is included in the name to emphasize how self-assembly can be simulated here in generalized, nonequilibrium systems such as the cell. Additionally, although the RD model is commonly applied to studying systems not at thermodynamic equilibrium, it is nonetheless useful for quantifying the pathways to and composition of equilibrium for complex multicomponent systems. The standard computational approach for studying self-assembly, using coarse-grained molecular modeling, provides access to these pathways and equilibria with more physically detailed models, in which interactions emerge because of distance-dependent energy functions rather than the rate-controlled events of RD models. A coarse molecular model thus naturally can accommodate a range of structures or defects and capture emergent cooperativity, strain, and localization of components. However, they are still relatively limited in the length and timescales of dynamics they can access, which does not extend to the cell scale. They are parameterized by potential energy functions, and converting these to the binding free energies or rate constants for comparison to experiment is nontrivial. Lastly, chemical reactions require covalent bond breaking that is not typically accessible in molecular modeling, preventing systematic and transferable methods for involving enzymatic or ATP-driven reactions ubiquitous in cells. These models are unable to simulate in vivo cell signaling, cytoskeletal dynamics, or clathrin-mediated endocytosis, for example. Our NERDSS software addresses this substantial application gap, and although it lacks the detail of energy-function based models, it is able to uniquely preserve important features of molecular assembly.
What is ioNERDSS?
ioNERDSS is a python package developed by our group to assist NERDSS simulations. Its provides user-friendly analysis tool for setting up models and analyzing output from the reaction-diffusion simulator NERDSS.
Who are we?
NERDSS was developed by the Johnson lab at the Thomas C. Jenkins Department of Biophysics at the Johns Hopkins University. Our multidisciplinary research group studies dynamical systems in biology. We address how macromolecular self-assembly is controlled to occur at the right place and time, with focus on endocytosis, viral exit, and transcriptional regulation. Group members are interested both in principles and design of self-assembly, as well as biological function in cells. They develop and apply theory, modeling, and simulations to these problems, with frequent collaboration with experimentalists.
